The Power of Predictive AI Models: Revolutionizing Cancer Research
Powerful computational tools, such as predictive artificial intelligence (AI) models, are reshaping preclinical research, leading to more optimized and streamlined drug discovery and drug development pipelines.1 In drug discovery, AI can be applied to identifying and validating relevant cancer biomarkers, which may turn into new drug targets or prognostic tools. In drug development, AI models are being generated to rapidly perform in silico tests on lead compounds with combination cancer therapies to predict efficacy and side effects.1,2
In the following article, we’ll look at what predictive AI is, the advantages of applying it in preclinical cancer research, some of its limitations, and how it has the potential to revolutionize translational oncology research.
The Current Challenges in Cancer Research
Preclinical cancer research is deepening researchers’ understanding of the biology of cancer, what makes a promising drug target, and how predictive biomarkers can reveal clinical outcomes. Unfortunately, there are still significant knowledge gaps that ultimately become translation gaps: Fewer than 4% of anti-cancer drug candidates that enter phase I clinical trials go on to be approved for use in patients.3
The reason for such low translation rates is multi-faceted. Cancer is complex, and answering preclinical and clinical questions relies on solving some prototypical challenges, including selecting the appropriate molecular target, indication, in vitro or in vivo model, biomarker, and cancer therapeutic combinations.
Navigating these choices can make or break future clinical success. Better choices in the preclinical phase of research – fueled by generating more clinically-relevant data and analyzing this data with state-of-the-art tools – can drive improved clinical translation.
With the promise of predictive AI in advancing cancer research and the rapidly evolving computational landscape, let's first define exactly what we mean when we talk about AI.
Defining AI Terminology
AI is a technology that mimics human intelligence, enabling a machine to learn and recognize patterns and relationships when given representative examples and to use what it learns to make predictions and/or decisions.1
representative examples and to use what it learns to make predictions and/or decisions.1
While machine learning (ML) algorithms are often used synonymously with AI, ML is a branch of AI that can learn and adapt based on training with structured data sets to predict outcomes or discover patterns in data.2
Deep learning is a subset of ML that uses multilayer neural networks loosely modeled on the organization of the human brain.4 Deep learning can handle problems that are difficult to define precisely using unstructured data (e.g., pictures, audio, etc.), such as categorizing images of skin lesions as benign or malignant.2,5 Convolutional neural networks (CNNs) are deep learning architectures that learn relevant features automatically and do not use manual curation like in traditional machine learning.
AI Applications in Cancer Research
The availability of extensive imaging, genomics, transcriptomics, and other ‘omics datasets in collections such as The Cancer Genome Atlas (TCGA) or The Pan-Cancer Analysis of Whole Genomes (PCAWG) has provided a solid foundation for predictive AI model development. AI systems need data for proper, unbiased training and validation, and these publicly-available data collections are great resources for advancing predictive AI in translational oncology.
In addition, the robust activity in cancer research provides a continuous stream of data to validate predictions generated from AI algorithms, leading to continued training and improved algorithm accuracy. Accordingly, several typical preclinical applications for AI have emerged.
Cancer Biomarker Identification
Identifying genetic variants from next-generation sequencing (NGS) data has become an integral technique for cancer diagnosis and predicting cancer treatment responses. Yet, it comes with a number of analytical challenges and only provides a static snapshot of the biomarkers present at the time of cancer biopsy.
One of the first successful applications of AI in cancer research was the development of DeepVariant, which solved several issues in NGS sequence analysis (e.g., low coverage, repeat regions, etc.) and enabled more accurate variant calling.6 Another application of AI in the biomarker space is predicting clinically-relevant mutations using imaging data (e.g., histopathology, radiology, etc.). Based on available image data, several studies have focused on developing algorithms that predict key driver mutations for specific cancer subtypes. Wang et al., for instance, were able to determine EGFR mutation status based on computed tomography (CT) images from over 800 lung adenocarcinoma patients.7 Other similar approaches have made predictions about microsatellite instability (MSI) status and tumor mutation burden (TMB) based on a variety of image types from diverse cancer types.1
Biomarkers (genetic or otherwise) are well-established for differentiating cancer patient groups that may experience metastases, recurrence, or treatment resistance, thus, making AI useful for choosing clinically relevant in vitro or in vivo cancer models. Several studies have used genomics, transcriptomics, and/or proteomics data to predict efficacy in specific cell lines, with high sensitivity and specificity.8 Cortés-Ciriano et al. modeled a common in vitro efficacy endpoint, the 50% growth inhibition bioassay (GI50), to predict growth inhibition across many cancer cell lines and tissues.9
In clinical testing, these same algorithms can be used to develop cancer patient stratification strategies enabling a more informed clinical trial design with a higher probability of success.
Uncovering Cancer Therapies' Mechanism of Action
Often in drug discovery and development, an anti-cancer drug candidate’s mechanism of action (MoA) is not fully understood. This knowledge gap can remain through clinical testing and even after approval. However, understanding an anti-cancer drug candidate’s MoA can help determine which preclinical cancer models to use and what synergies with other cancer therapies may exist.
AI algorithms have helped predict cancer drug MoAs. A deep learning model named DrugCell was trained on the response of 1,235 distinct tumor cell lines to 684 different anti-cancer drugs. Based on chemical structure, the AI model could predict drug response, underlying MoA, and synergistic cancer therapy combinations.10
Identifying Synergistic Cancer Drug Combination Selection
Combining multiple synergistic anti-cancer drugs can overcome drug resistance to targeted therapy. Machine learning has been used to predict drug response and rational combination therapy based on dynamic signaling responses in cancer cells from individuals with lung cancer exposed ex vivo to targeted anticancer drugs. Such approaches were able to guide clinical treatment decisions.11
Experimental and Predicted Cancer Drug Combinations11
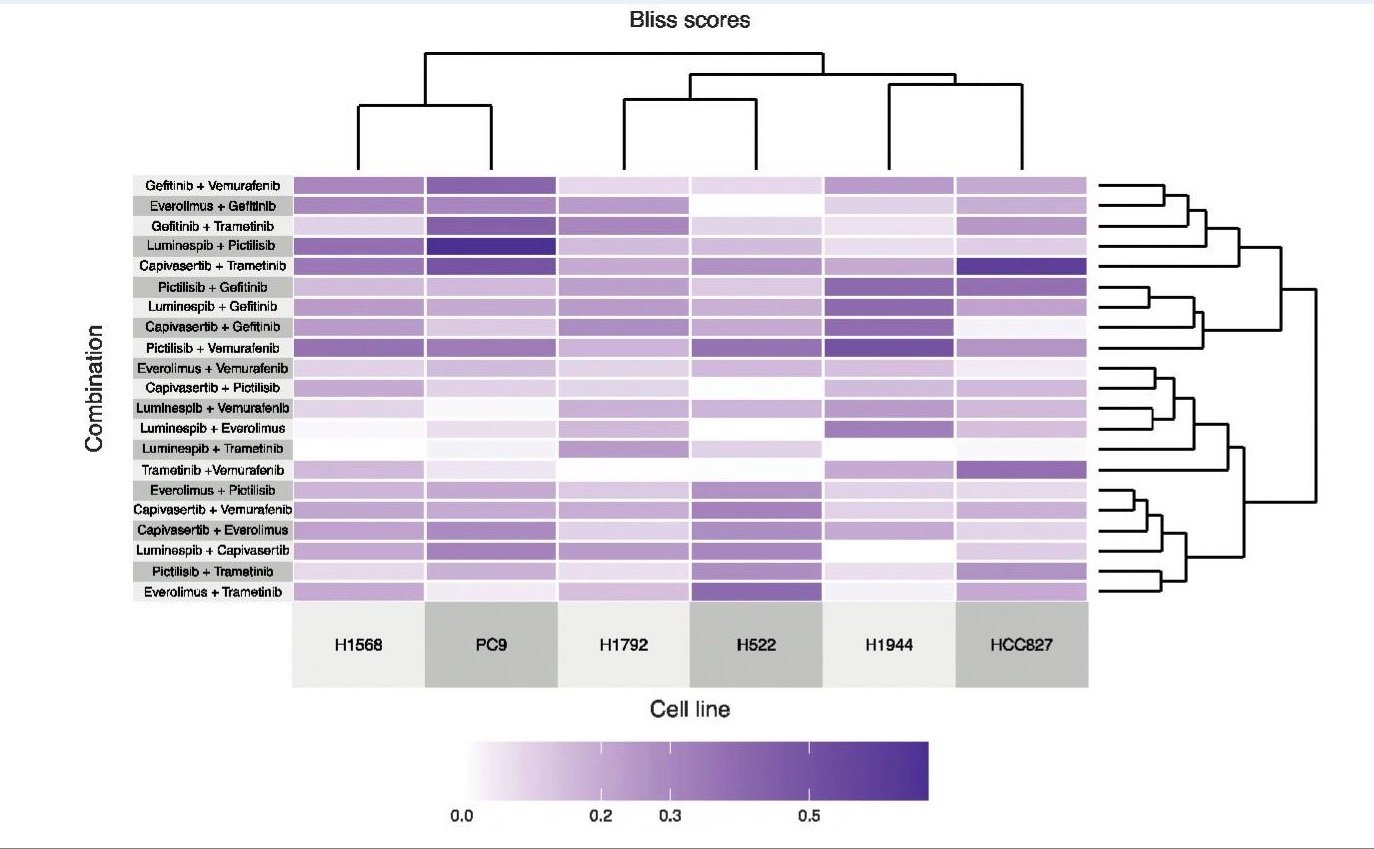
Figure 1: Clustered heatmap of Bliss synergy scores was experimentally measured for six cancer cell lines treated with 21 two-drug combinations.11
Additionally, the AstraZeneca-Sanger Drug Combination Prediction DREAM Challenge explored ML approaches for predicting synergistic anti-cancer drug combinations at preclinical stages.12
Anti-Cancer Drug Combinations and Cancer Cell Lines Profiled12
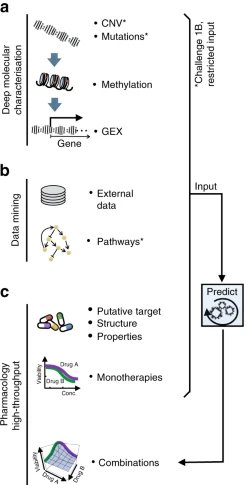
Figure 2: a Molecular characterization of the cancer cell lines included genetics, epigenetics, and transcriptomics. b Participants were encouraged to mine external data and pathway resources. c Participants were provided the putative targets for all and chemical structures for ~⅓ of drugs (with this manuscript structures are now provided for all drugs).12
Cancer Indication Selection
Choosing the most promising indication for a new anti-cancer drug is critical in therapeutic development. AI solutions, such as the PREDICT algorithm, have been developed that use drug-drug and disease-disease similarity score datasets to predict potential indications for novel drugs.13 PREDICT was trained on a large drug-disease association dataset and can also be used for predicting new indications for existing, approved anti-cancer drugs (e.g., drug repurposing), reducing the time and cost of drug development.
Transcriptome data can also be used to identify anti-cancer drugs for repurposing: Transcriptional data from the Library of Integrated Network-Based Cellular Signatures (LINCS), containing gene perturbation profiles, have been used to train deep neural networks. This approach identified repurposing drug candidates that can reverse expression profiles of cancer-specific gene signatures in bladder, colorectal, and liver cancer.14–16
Repurposing Drugs Targeting Co-Upregulated/Co-Downregulated Genes in Colorectal Cancer15
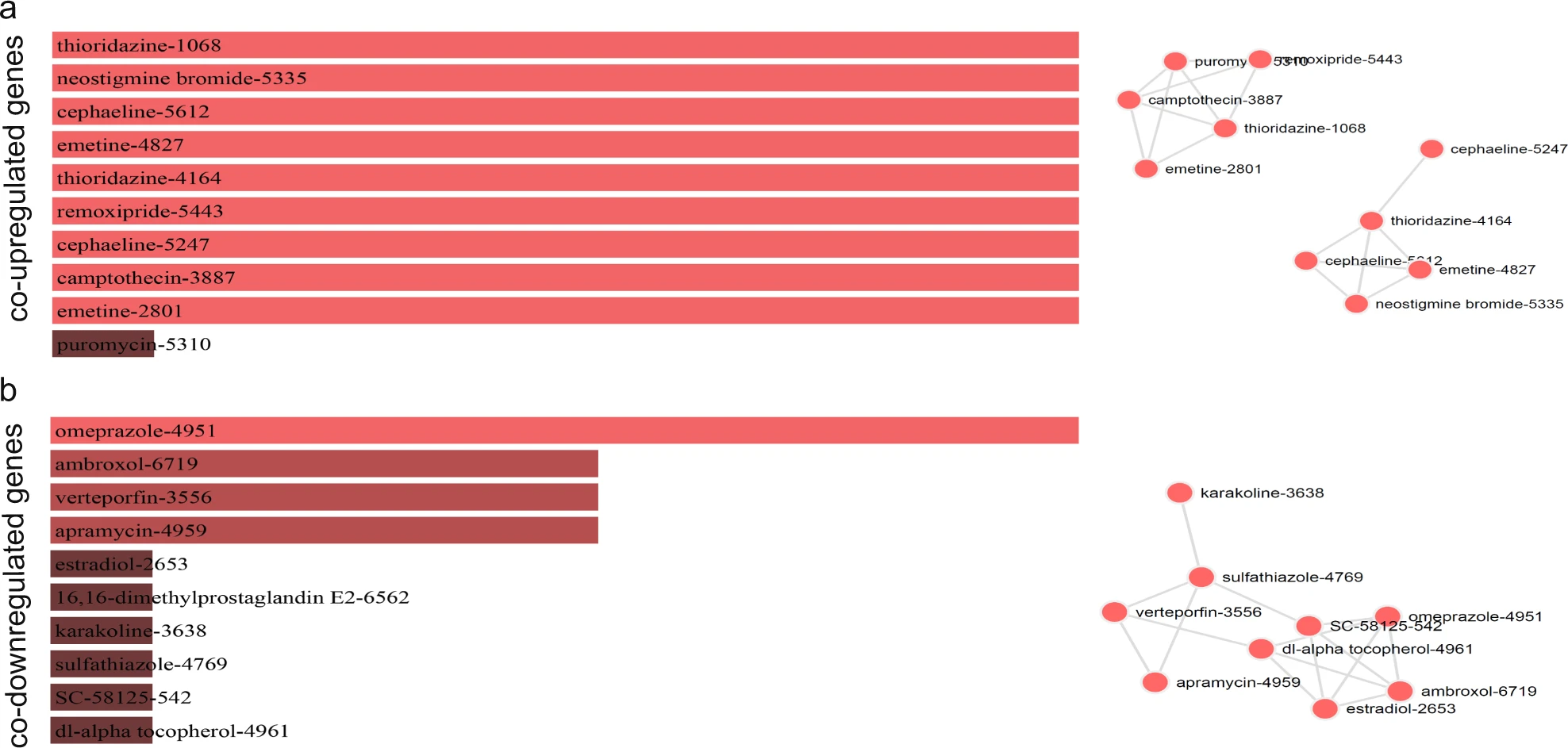
Figure 3: Τhe bar graphs are sorted by the combined score. The length of each bar represents the significance of its corresponding term. The brighter the color, the more significant that term is. The drugs in the network are sized according to their degree (number of edges), whereas the thickness of a connecting edge is proportional to the partial correlation coefficient between the two drugs. The nodes are arranged so that the edges are of more or less equal length and there are as few edge crossings as possible. For clarity, only the top 10 drugs ranked by partial correlation coefficient are shown.15
These AI-driven tools (mentioned in the above sections) can prioritize anti-cancer drug candidates based on user-selected parameters and priorities.
Advantages of AI in Cancer Research
As described above, using AI in cancer research may bring several benefits to drug developers:
- Cost savings: Drug development is costly, with large amounts of investment required for preclinical studies, clinical trials, and manufacturing. Failures in any of these areas lead to a loss of time and money for developers and an increase in the overall cost of bringing a drug to market. Using AI helps to avoid these failures by predicting efficacious drugs/drug combinations, which cancer models are best for preclinical testing, and which patients to focus clinical testing on.
- More precise precision oncology: AI can help identify the genetic and molecular biomarkers of individual tumors and which are most predictive of specific clinical outcomes (e.g., treatment response, recurrence, metastasis, etc.). Ultimately, AI applications in cancer research have the potential to usher in a new era of personalized oncology.
- More focused validation: In silico algorithms have been shown to be highly predictive is powerful and in vitro, in vivo, and clinical studies act as complementary methods to experimentally validate results from AI algorithms.1 AI can be used to provide a more focused, precise, and iterative approach to preclinical testing. AI enables informed decision-making about which in vitro and in vivo models to use. The resulting data has two core benefits: data can be used to 1) further refine AI-predictive models and 2) choose different in vitro and in vivo cancer models to advance preclinical testing.
Limitations of AI in Cancer Research
The AI field has been around for a long time. In 1956, at a now famous Dartmouth summer conference, the term artificial intelligence is used for the first time and leaders in the field launched AI research as a legitimate area of focus.17 While much has been accomplished since then, including the recent popularization of large language models, such as ChatGPT and others, recent life science advances and applications have raised many more questions and challenges for AI researchers to address.
Thus, AI is not a cure-all. There are some current limitations, including:
- Data bias and quality: Data is what drives advances in AI. If the data used for training and validating AI systems is biased (e.g., focused on one cancer type, patient population, primary vs. metastatic tumors, etc.) or low quality (e.g., small, statistically-insignificant), this can negatively impact the predictive sensitivity and selectivity. Some AI applications mentioned above rely heavily on data sets generated using cell cultures rather than more clinically relevant patient-derived models, such as, 3D tumor spheroids or patient-derived xenografts (PDX). Data from cancer models that mimic the clinical progression of cancer will help develop AI algorithms that translate to the clinic, bridging the current translation gap.
- Interpretability: Predictive AI models are often called black boxes because of an inability to see or interpret which biological mechanisms were used to drive the prediction.10 This limitation has long been appreciated, but only recently has a focus on developing methods around interpretability come about. DrugCell is a great example of an interpretable deep learning model that allows users to see the underlying mechanism used to predict response to therapy. As AI continues to be applied across many industries, interpretability will surely increase, making AI-generated insights more accurate, meaningful, and, importantly, transparent.
- Cost: While AI can make preclinical and clinical cancer research programs more cost-efficient in the long run, implementing an AI system can provide a hefty short-term investment, which may make it cost-prohibitive for smaller organizations. There are costs to construct and maintain a computational infrastructure, train and validate an AI model, and hire experienced computer scientists or bioinformaticians. The cost of implementing AI in preclinical cancer research is likely to decrease over time as the technology becomes more widely adopted and the costs of infrastructure, training, and expertise decrease. Another option for anti-cancer drug developers is to outsource AI expertise to the increasing number of off-the-shelf and custom solutions that have become available through specialized contract research organizations (CROs).
The Future of AI in Cancer Research
AI will continue to transform preclinical and clinical cancer research. The future of AI in preclinical and clinical cancer research may be characterized by increased efficiency, improved accuracy, and more personalized cancer treatments. These advancements will be driven by developing more advanced, predictive AI models and training using more clinically-relevant data from PDX and other patient-derived cancer models. While challenges remain, including the need for more robust data and improved interpretability of AI-generated insights, the potential benefits of AI in preclinical cancer research are significant and hold great promise for improving patient outcomes in the fight against cancer.
CertisAI™: Bridging the Translation Gap with AI-Predictive Insights
Certis Oncology Solutions is the only translational science partner that combines the predictive power of AI and deep expertise in cancer model development to reliably answer complex questions about therapeutic effects. CertisAI Predictive Oncology Intelligence™ uses multivariate machine learning algorithms to capture the nuance of biomarker interactions and bring AI-enabled accuracy to cancer model selection, predictions of drug efficacy, and biomarker identification. Its proprietary in silico platform utilizes big data, statistical algorithms, and machine learning to predict anti-cancer drug efficacy based on gene expression biomarkers. This pan-cancer solution can accelerate drug discovery and companion diagnostics development.
CertisAI integrates with Certis’ deep experience in the custom development of orthotopic PDX (O-PDX) models, which are used to validate in silico predictions.
Learn more about leveraging data science's power for reproducible, actionable preclinical results.
Schedule a Meeting Watch a Webinar View Press Release Request Info
References
- Bhinder B, Gilvary C, Madhukar NS, Elemento O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. (2021) 11, 900-915.
- Bernstam EV, Shireman PK, Meric-Bernstam F, et al. Artificial Intelligence in Clinical and Translational Science: Successes, Challenges and Opportunities. Clin Transl Sci. (2022) 15, 309-321.
- Wong CH, Siah KW, Lo AW. Estimation of Clinical Trial Success Rates and Related Parameters. Biostat Oxf Engl. (2019) 20, 273-286.
- Study Urges Caution when Comparing Neural Networks to the Brain. MIT News website. Published November 2, 2022. Accessed May 8, 2023.
- Liang G, Fan W, Luo H, Zhu X. The Emerging Roles of Artificial Intelligence in Cancer Drug Development and Precision Therapy. Biomed Pharmacother. (2020) 128, 110255.
- Poplin R, Chang PC, Alexander D, et al. A Universal SNP and Small-Indel Variant Caller using Deep Neural Networks. Nat Biotechnol. (2018) 36, 983-987.
- Wang S, Shi J, Ye Z, et al. Predicting EGFR Mutation Status in Lung Adenocarcinoma on Computed Tomography Image Using Deep Learning. Eur Respir J. (2019) 53, 1800986.
- Ding MQ, Chen L, Cooper GF, Young JD, Lu X. Precision Oncology beyond Targeted Therapy: Combining Omics Data with Machine Learning Matches the Majority of Cancer Cells to Effective Therapeutics. Mol Cancer Res. (2018) 16, 269-278.
- Cortés-Ciriano I, van Westen GJP, Bouvier G, et al. Improved Large-Scale Prediction of Growth Inhibition Patterns Using the NCI60 Cancer Cell Line Panel. Bioinformatics. (2016) 32, 85-95.
- Kuenzi BM, Park J, Fong SH, et al. Predicting Drug Response and Synergy Using a Deep Learning Model of Human Cancer Cells. Cancer Cell. (2020) 38, 672-684.e6.
- Coker EA, Stewart A, Ozer B, et al. Individualized Prediction of Drug Response and Rational Combination Therapy in NSCLC Using Artificial Intelligence–Enabled Studies of Acute Phosphoproteomic Changes. Mol Cancer Ther. (2022) 21, 1020-1029.
- Menden MP, Wang D, Mason MJ, et al. Community Assessment to Advance Computational Prediction of Cancer Drug Combinations in a Pharmacogenomic Screen. Nat Commun. (2019) 10, 2674.
- Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: A Method for Inferring Novel Drug Indications with Application to Personalized Medicine. Mol Syst Biol. 2011 7, 496.
- Chen B, Ma L, Paik H, et al. Reversal of Cancer Gene Expression Correlates with Drug Efficacy and Reveals Therapeutic Targets. Nat Commun. (2017) 8, 16022.
- Mastrogamvraki N, Zaravinos A. Signatures of Co-Deregulated Genes and Their Transcriptional Regulators in Colorectal Cancer. Npj Syst Biol Appl. (2020) 6, 1-16.
- Mokou M, Lygirou V, Angelioudaki I, et al. A Novel Pipeline for Drug Repurposing for Bladder Cancer Based on Patients’ Omics Signatures. Cancers. (2020) 12, 3519.
- The History of Artificial Intelligence. Science in the News website. Published August 28, 2017. Accessed May 25, 2023.


